Neoadjuvant immunochemotherapy may offer superior outcomes compared to concurrent chemoradiotherapy (CCRT) in patients with limited-stage small cell lung cancer (LS-SCLC), according to a new report.
The data suggests immunochemotherapy generates…
Neoadjuvant immunochemotherapy may offer superior outcomes compared to concurrent chemoradiotherapy (CCRT) in patients with limited-stage small cell lung cancer (LS-SCLC), according to a new report.
The data suggests immunochemotherapy generates…

Ladha JK, Jat ML, Stirling CM, Chakraborty D, Pradhan P, Krupnik TJ et al. Achieving the sustainable development goals in agriculture: The crucial role of nitrogen in cereal-based systems. In: Sparks DL, editor. Advances in Agronomy, Vol 163. Advances in Agronomy. 1632020. pp. 39–116.
Menegat S, Ledo A, Tirado R. Greenhouse gas emissions from global production and use of nitrogen synthetic fertilisers in agriculture. Sci Rep. 2022;12(1):14490.
Chauhan BSJ, Khawar J, Mahajan G. Rice production worldwide. Springer. 2017;247.
Xu JZ, Peng SZ, Yang SH, Wang WG. Ammonia volatilization losses from a rice paddy with different irrigation and nitrogen managements. Agric Water Manage. 2012;104:184–92.
Google Scholar
Hu B, Wang W, Chen JJ, Liu YQ, Chu CC. Genetic improvement toward nitrogen-use efficiency in rice: lessons and perspectives. Mol Plant. 2023;16(1):64–74.
Google Scholar
Neeraja CN, Barbadikar KM, Mangrauthia SK, Rao PR, Subrahmanayam D, Sundaram RM. Genes for NUE in rice: a way forward for molecular breeding and genome editing. Plant Physiol Rep. 2021;26(4):587–99.
Google Scholar
Cassman KG, Peng S, Olk DC, Ladha JK, Reichardt W, Dobermann A, et al. Opportunities for increased nitrogen-use efficiency from improved resource management in irrigated rice systems. Field Crops Res. 1998;56(1–2):7–39.
Google Scholar
Li H, Hu B, Chu CC. Nitrogen use efficiency in crops: lessons from Arabidopsis and rice. J Exp Bot. 2017;68(10):2477–88.
Google Scholar
Reig-Valiente J, Viruel J, Sales E, Marqués L, Terol J, Gut M et al. Genetic diversity and population structure of rice varieties cultivated in temperate regions. Rice. 2016;9:58.
Andaya VC, Tai TH. Fine mapping of the qCTS12 locus, a major QTL for seedling cold tolerance in rice. Theor Appl Genet. 2006;113(3):467–75.
Google Scholar
Hour AL, Hsieh WH, Chang SH, Wu YP, Chin HS, Lin YR. Genetic diversity of landraces and improved varieties of rice (Oryza sativa L.) in Taiwan. Rice. 2020;13:82.
Kumar V, Rithesh L, Raghuvanshi N, Kumar A, Parmar K. Advancing nitrogen use efficiency in cereal crops: A comprehensive exploration of genetic manipulation, nitrogen dynamics, and plant nitrogen assimilation. South Afr J Bot. 2024;169:486–98.
Google Scholar
Nazish T, Arshad M, Jan SU, Javaid A, Khan MH, Naeem MA, et al. Transporters and transcription factors gene families involved in improving nitrogen use efficiency (NUE) and assimilation in rice (Oryza sativa L). Transgenic Res. 2022;31(1):23–42.
Google Scholar
Hou MM, Yu M, Li ZQ, Ai ZY, Chen JG. Molecular regulatory networks for improving nitrogen use efficiency in rice. Int J Mol Sci. 2021;22(16):9040.
Chaudhary S, Kalkal M. Rice transcriptome analysis reveals nitrogen starvation modulates differential alternative splicing and transcript usage in various Metabolism-Related genes. Life-Basel. 2021;11(4):285.
Feng Y, Lu Q, Zhai RR, Zhang MC, Xu Q, Yang YL, et al. Genome wide association mapping for grain shape traits in indica rice. Planta. 2016;244(4):819–30.
Google Scholar
Qiu XJ, Zhu SB, Hu H, Wang CC, Lv WK, He LP, et al. Genome-Wide association mapping for grain shape in rice accessions. Int J Agric Biology. 2020;23(3):582–8.
Google Scholar
Liu YZ, Xin W, Chen LQ, Liu YQ, Wang X, Ma C et al. Genome-Wide association analysis of effective tillers in rice under different nitrogen gradients. Int J Mol Sci. 2024;25(5):2969.
Cheng XP, Chang YP, Sun JH, Du MY, Liang LP, Zhang MY et al. Identification of candidate genes and favourable haplotypes for yield traits in rice based on a genome-wide association study. Euphytica. 2023;219(12):125.
Peng SS, Liu YC, Xu YC, Zhao JH, Gao P, Liu Q et al. Genome-Wide association study identifies a Plant-Height-Associated Gene OsPG3 in a population of commercial rice varieties. Int J Mol Sci. 2023;24(14):11454.
Rakotoson T, Dusserre J, Letourmy P, Frouin J, Ratsimiala IR, Rakotoarisoa NV, et al. Genome-Wide association study of nitrogen use efficiency and agronomic traits in upland rice. Rice Sci. 2021;28(4):379–90.
Google Scholar
Wang YY, Cheng YH, Chen KE, Tsay YF. Nitrate Transport, Signaling, and Use Efficiency. In: Merchant SS, editor. Annual Review of Plant Biology, Vol 69. Annual Review of Plant Biology. 692018. pp. 85–122.
Wei D, Cui KH, Ye GY, Pan JF, Xiang J, Huang JL, et al. QTL mapping for nitrogen-use efficiency and nitrogen-deficiency tolerance traits in rice. Plant Soil. 2012;359(1–2):281–95.
Google Scholar
Li Q, Lu XL, Wang CJ, Shen L, Dai LP, He JL, et al. Genome-wide association study and transcriptome analysis reveal new QTL and candidate genes for nitrogen-deficiency tolerance in rice. Crop J. 2022;10(4):942–51.
Google Scholar
Ding CQ, You J, Chen L, Wang SH, Ding YF. Nitrogen fertilizer increases spikelet number per panicle by enhancing cytokinin synthesis in rice. Plant Cell Rep. 2014;33(2):363–71.
Google Scholar
Das K, Panda BB, Shaw BP, Das SR, Dash SK, Kariali E et al. Grain density and its impact on grain filling characteristic of rice: mechanistic testing of the concept in genetically related cultivars. Sci Rep. 2018;8:4149.
Gao HB, Wang WG, Wang YH, Liang Y. Molecular mechanisms underlying plant architecture and its environmental plasticity in rice. Mol Breeding. 2019;39(12):167.
Dou Z, Zhou YC, Zhang YY, Guo W, Xu Q, Gao H. Influence of nitrogen applications during grain-Filling stage on rice (Oryza sativa L.) yield and grain quality under high temperature. Agronomy-Basel. 2024;14(1):216.
Li RH, Li MJ, Ashraf U, Liu SW, Zhang JE. Exploring the relationships between yield and yield-Related traits for rice varieties released in China from 1978 to 2017. Front Plant Sci. 2019;10:543.
Zhang QG, Sun J, Wang LP, Chen J, Ke J, Wu LQ. Effects of nitrogen application at different panicle development stages on the panicle structure and grain yield in hybrid indica rice cultivars. Agronomy-Basel. 2025;15(3):595.
Xin W, Wang JG, Li J, Zhao HW, Liu HL, Zheng HL et al. Candidate gene analysis for nitrogen absorption and utilization in Japonica rice at the seedling stage based on a Genome-Wide association study. Front Plant Sci. 2021;12:670861.
García-Romeral J, Castanera R, Casacuberta J, Domingo C. Deciphering the genetic basis of allelopathy in japonica rice cultivated in temperate regions using a Genome-Wide association study. Rice. 2024;17(1):22.
Walkley A, Black IA. An examination of the Degtjareff method for determining soil organic matter, and a proposed modification of the chromic acid titration method. Soil Sci. 1934;37(1):29–38.
Google Scholar
Bouyoucos GJ. The hydrometer as a new method for the mechanical analysis of soils. Soil Sci. 1927;23(5):343–54.
Google Scholar
Lancashire PD, Bleiholder H, Vandenboom T, Langeluddeke P, Stauss R, Weber E, et al. A uniform decimal code for growth-stages of crops and weeds. Ann Appl Biol. 1991;119(3):561–601.
Google Scholar
Inc. SI. JMP®. 16 ed. Cary, NC: SAS Institute Inc.; 1989–2023.
Team RC. R: A Language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2023.
Kowarik A, Templ M. Imputation with the R package VIM. J Stat Softw. 2016;74(7):1–16.
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23(19):2633–5.
Google Scholar
Yu J, Pressoir G, Briggs WH, Vroh Bi I, Yamasaki M, Doebley JF, et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet. 2006;38(2):203–8.
Google Scholar
Patil I. Visualizations with statistical details: the ‘ggstatsplot’ approach. J Open Source Softw. 2021;6:3167.
Google Scholar
Sakai H, Lee SS, Tanaka T, Numa H, Kim J, Kawahara Y, et al. Rice annotation project database (RAP-DB): an integrative and interactive database for rice genomics. Plant Cell Physiol. 2013;54(2):e6.
Google Scholar
Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Volume 6. New York, NY: Rice; 2013. p. 4. 1.
McCouch SR, Rice Genetics C. Gene nomenclature system for rice. Rice. 2008;1(1):72–84.
Google Scholar
Kurata N, Yamazaki Y, Oryzabase. An integrated biological and genome information database for rice. Plant Physiol. 2006;140(1):12–7.
Google Scholar
Mansueto L, Fuentes RR, Borja FN, Detras J, Abriol-Santos JM, Chebotarov D, et al. Rice SNP-seek database update: new snps, indels, and queries. Nucleic Acids Res. 2017;45(D1):D1075–81.
Google Scholar
Alexandrov N, Tai SS, Wang WS, Mansueto L, Palis K, Fuentes RR, et al. SNP-Seek database of SNPs derived from 3000 rice genomes. Nucleic Acids Res. 2015;43(D1):D1023–7.
Google Scholar
Yang WZ, Yoon J, Choi H, Fan YL, Chen RM, An G. Transcriptome analysis of nitrogen-starvation-responsive genes in rice. BMC Plant Biol. 2015;15:31.
Yan M, Fan XR, Feng HM, Miller AJ, Shen QR, Xu GH. Rice OsNAR2.1 interacts with OsNRT2.1, OsNRT2.2 and OsNRT2.3a nitrate transporters to provide uptake over high and low concentration ranges. Plant Cell Environ. 2011;34(8):1360–72.
Google Scholar
Liu XQ, Huang DM, Tao JY, Miller AJ, Fan XR, Xu GH. Identification and functional assay of the interaction motifs in the partner protein OsNAR2.1 of the two-component system for high-affinity nitrate transport. New Phytol. 2014;204(1):74–80.
Google Scholar
Jiang HZ, Wang YM, Lai LR, Liu XT, Miao CJ, Liu RF, et al. OsAMT1.1 Expression by Nitrate-Inducible promoter of OsNAR2.1 Increases nitrogen use efficiency and rice yield. Rice Sci. 2023;30(3):222–34.
Google Scholar
Chen HL, Xu N, Wu Q, Yu B, Chu YL, Li XX, et al. OsMADS27 regulates the root development in a NO3-dependent manner and modulates the salt tolerance in rice (Oryza sativa L.). Plant Sci. 2018;277:20–32.
Google Scholar
Pachamuthu K, Sundar VH, Narjala A, Singh RR, Das S, Pal H, et al. Nitrate-dependent regulation of miR444-OsMADS27 signalling cascade controls root development in rice. J Exp Bot. 2022;73(11):3511–30.
Google Scholar
Singh RK, Khan W, editors. Effect of nitrogen levels on growth and yield of basmati rice varieties2021.
Li G, Zhang Y, Zhou C, Xu J, Cj Z, Ni C, et al. Agronomic and physiological characteristics of high yield and nitrogen use efficient varieties of rice: comparison between two near-isogenic lines. Food Energy Secur. 2024;13(2):e539.
Google Scholar
Tayefe M, Gerayzade A, Amiri E, Zade AN. Effect of nitrogen on rice yield, yield components and quality parameters. 2014.
Haghshenas H, Ghanbari Malidarreh A. Response of yield and yield components of released rice cultivars from 1990–2010 to nitrogen rates. Cent Asian J Plant Sci Innov. 2021;1(1):23–31.
Zhang SY, Huang YZ, Ji Z, Fang YZ, Tian YN, Shen CB, et al. Discovery of SOD5 as a novel regulator of nitrogen-use efficiency and grain yield via altering auxin level. New Phytol. 2025;246(3):1084–95.
Google Scholar
Wu J, Sun LQ, Song Y, Bai Y, Wan GY, Wang JX, et al. The OsNLP3/4-OsRFL module regulates nitrogen-promoted panicle architecture in rice. New Phytol. 2023;240(6):2404–18.
Google Scholar
Duan YH, Zhang YL, Ye LT, Fan XR, Xu GH, Shen QR. Responses of rice cultivars with different nitrogen use efficiency to partial nitrate nutrition. Ann Botany. 2007;99(6):1153–60.
Google Scholar
Tsednee M. Linking timing to nitrogen use efficiency: rice OsEC-Ghd7-ARE1 module works on it. Plant Physiol. 2024;196(3):1720–1.
Google Scholar
Zhang L, Shen C, Zhu S, Ren N, Chen K, Xu J. Effects of sowing date and nitrogen (N) application rate on grain yield, nitrogen use efficiency and 2-acetyl-1-pyrroline formation in fragrant rice. Agronomy. 2022;12(12):3035.
Google Scholar
Zhang SN, Zhang YY, Li KN, Yan M, Zhang JF, Yu M, et al. Nitrogen mediates flowering time and nitrogen use efficiency via floral regulators in rice. Curr Biol. 2021;31(4):671–83.
Google Scholar
Mo Y, Jeong JM, Ha SK, Kim J, Lee C, Lee GP et al. Characterization of QTLs and candidate genes for days to heading in rice Recombinant inbred lines. Genes. 2020;11(9):957.
Song J, Tang LQ, Cui YT, Fan HH, Zhen XQ, Wang JJ. Research progress on photoperiod gene regulation of heading date in rice. Curr Issues Mol Biol. 2024;46(9):10299–311.
Google Scholar
Reig-Valiente JL, Marqués L, Talón M, Domingo C. Genome-wide association study of agronomic traits in rice cultivated in temperate regions. BMC Genomics. 2018;19:706.
Song YL, Gao ZC, Luan WJ. Interaction between temperature and photoperiod in regulation of flowering time in rice. Sci China-Life Sci. 2012;55(3):241–9.
Google Scholar
Matsubara K, Yamanouchi U, Wang ZX, Minobe Y, Izawa T, Yano M. Ehd2, a rice ortholog of the maize INDETERMINATE1 gene, promotes flowering by up-regulating Ehd1. Plant Physiol. 2008;148(3):1425–35.
Google Scholar
Ohkubo Y, Kuwata K, Matsubayashi Y. A type 2 C protein phosphatase activates high-affinity nitrate uptake by dephosphorylating NRT2.1. Nat Plants. 2021;7(3):310.
Google Scholar
Wu JY, Yang SQ, Chen NA, Jiang QN, Huang LL, Qi JX et al. Nuclear translocation of OsMADS25 facilitated by OsNAR2.1 in reponse to nitrate signals promotes rice root growth by targeting OsMADS27 and OsARF7. Plant Commun. 2023;4(6):100642.
Alfatih A, Zhang J, Song Y, Jan SU, Zhang ZS, Xia JQ et al. Nitrate-responsive OsMADS27 promotes salt tolerance in rice. Plant Commun. 2023;4(2):100458.
Song X, Xiong Y, Kong X, Huang G. Roles of auxin response factors in rice development and stress responses. Plant Cell Environ. 2023;46(4):1075–86.
Google Scholar
Zhang S, Wu T, Liu S, Liu X, Jiang L, Wan J. Disruption of OsARF19 is critical for floral organ development and plant architecture in rice (Oryza sativa L). Plant Mol Biology Report. 2016;34:748–60.
Google Scholar
Gho YS, Song MY, Bae DY, Choi H, Jung KH. Rice PIN auxin efflux carriers modulate the nitrogen response in a changing nitrogen growth environment. Int J Mol Sci. 2021;22(6):3243.
Zhang S, Zhu L, Shen C, Ji Z, Zhang H, Zhang T, et al. Natural allelic variation in a modulator of auxin homeostasis improves grain yield and nitrogen use efficiency in rice. Plant Cell. 2020;33(3):566–80.
Google Scholar
Zhang S, Wang S, Xu Y, Yu C, Shen C, Qian Q, et al. The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1. Plant Cell Environ. 2015;38(4):638–54.
Google Scholar
Wu D, Cao Y, Wang D, Zong G, Han K, Zhang W, et al. Auxin receptor OsTIR1 mediates auxin signaling during seed filling in rice. Plant Physiol. 2024;194(4):2434–48.
Google Scholar
Cortes LT, Zhang ZW, Yu JM. Status and prospects of genome-wide association studies in plants. Plant Genome. 2021;14(1):e20077.
Curtin SJ, Tiffin P, Guhlin J, Trujillo DI, Burghardt LT, Atkins P, et al. Validating genome-wide association candidates controlling quantitative variation in nodulation. Plant Physiol. 2017;173(2):921–31.
Google Scholar
COLOMBO, Oct. 29 (Xinhua) — The highest number of dengue mosquito breeding sites in Sri Lanka have been found within school premises, local media News First reported Monday, citing the National Dengue Control Unit (NDCU).
Prashila…

Duchess Meghan and Prince Harry are swapping royal blue for Dodgers blue.
The royal couple, who have been California-bound for the past several years, were spotted sitting just behind the net at Game 4 of the World Series. The match, between the…

DOWNERS GROVE, Ill., Oct. 29, 2025 (GLOBE NEWSWIRE) — Strengthening cybersecurity readiness by learning from past mistakes will highlight the conversation when CompTIA, the leading global provider of vendor-neutral information technology (IT)…

A new study has found that the benefits of a fatty acid known as eicosapentaenoic acid vary widely from person to person. Researchers say the results highlight how individual metabolism plays a crucial role in protecting against cardiovascular…
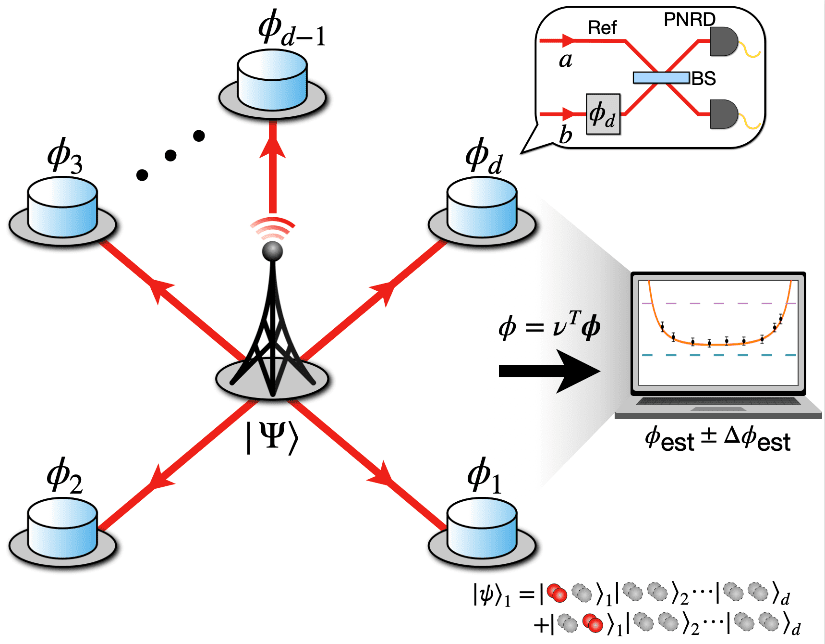
Hyang-Tag Lim’s research team at the Center for Quantum Technology, Korea Institute of Science and Technology (KIST), has demonstrated the world’s first ultra-high-resolution distributed quantum sensor network. By employing a special…

How will the reclassification of gasoline as carcinogenic to humans, impact dutyholders and their insurers?
The IARC has recently reclassified automative gasoline…

The emergency medical support team (EMST) was called to provide assistance at a home on the 9th floor in a 49-year-old Chinese man’s flat due to prolonged arm entrapment. His tenant discovered him on the bathroom floor with his right arm…
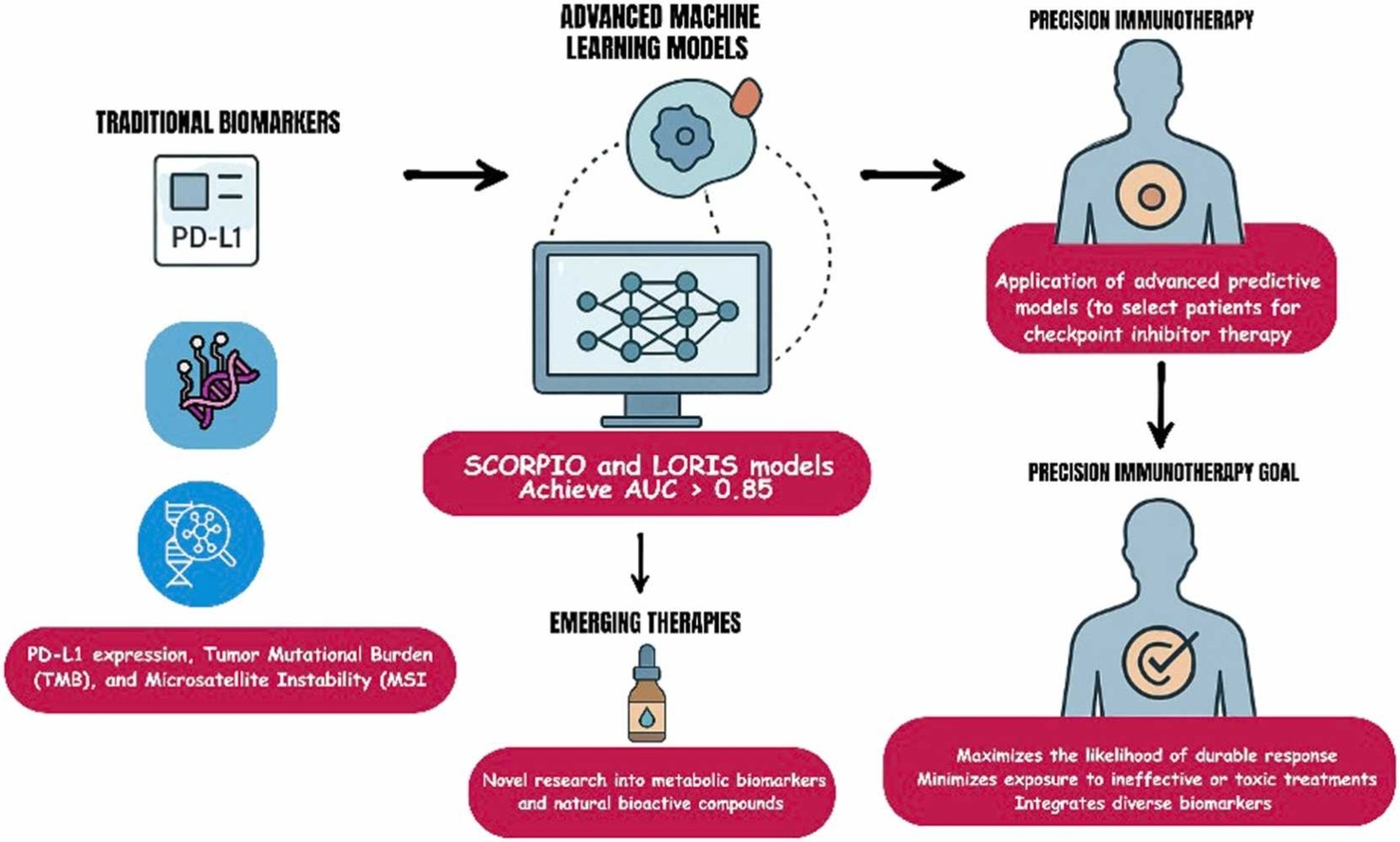
Immunotherapy, particularly immune checkpoint inhibition, has revolutionized modern oncology by achieving durable remissions across a wide range of tumor types. Despite these transformative outcomes, only 20–30% of patientsexperience sustained benefit, underscoring a critical challenge in accurately predicting who will respond to treatment. The clinical and economic implications are substantial—given that the global ICI market is projected to surpass $189 billion by 2032, the urgency for precise, clinically actionable predictive tools has never been greater.
In this comprehensive and forward-looking review, Oisakede et al. explore the rapidly advancing field of predictive modeling for immunotherapy response, encompassing biomarker-driven strategies, artificial intelligence and machine learning algorithms, mechanistic modeling, and multi-modal integration frameworks. Drawing from more than 200 studies, the authors deliver one of the most extensive comparative analyses to date, evaluating predictive accuracy, clinical applicability, and translational readiness across emerging methodologies.
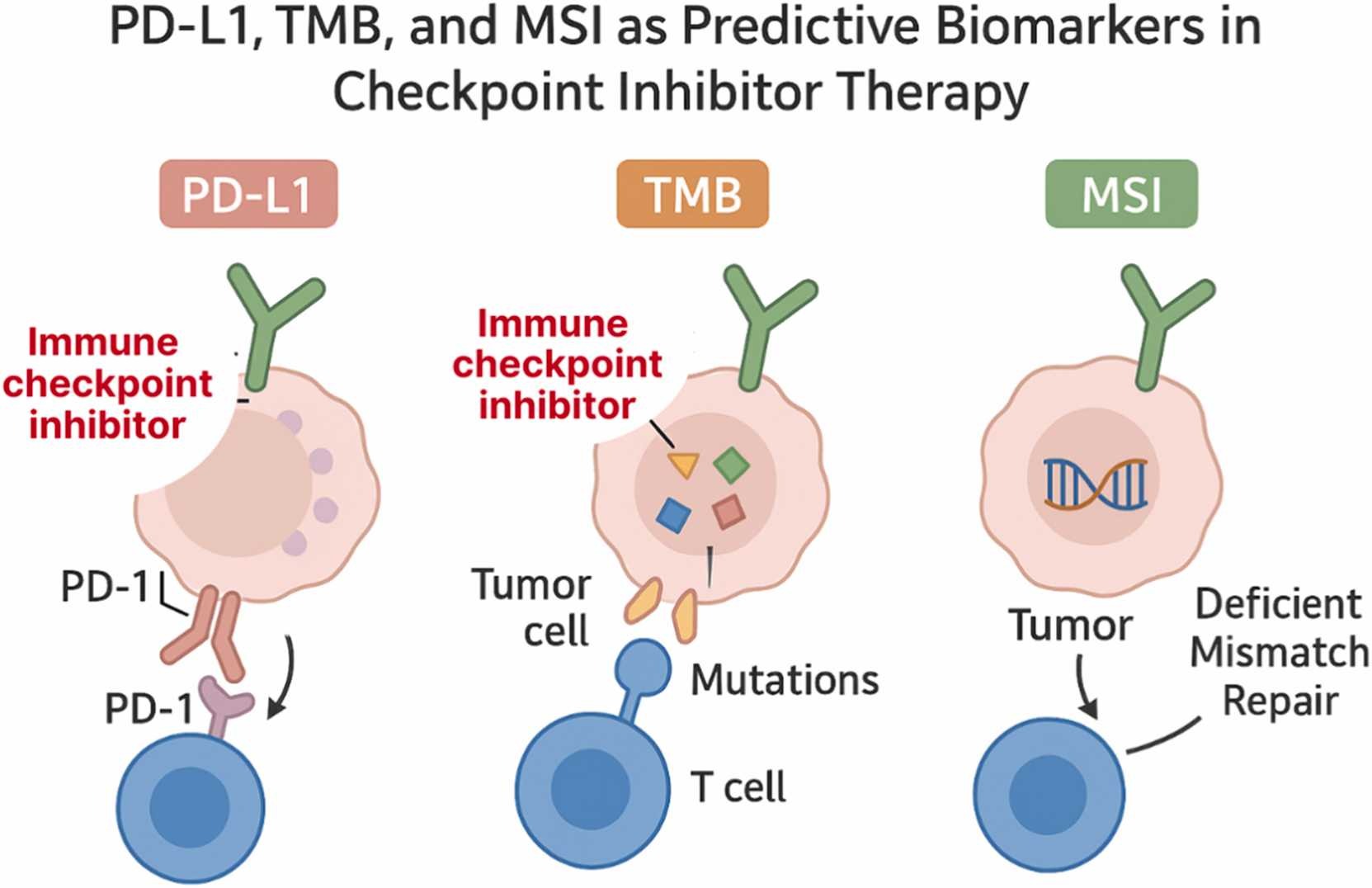
Traditional biomarkers—such as PD-L1 expression, tumor mutational burden (TMB), and microsatellite instability (MSI)—have long served as the foundation for selecting patients who might benefit from immune checkpoint inhibitors. However, their predictive accuracy remains limited.
PD-L1 expression, for instance, is predictive in only about 29% of FDA-approved indications, and both TMB and MSI show highly variable reliability across cancer types. These inconsistencies stem from biological heterogeneity, differences in testing platforms, and the complex interplay between tumor and immune factors.
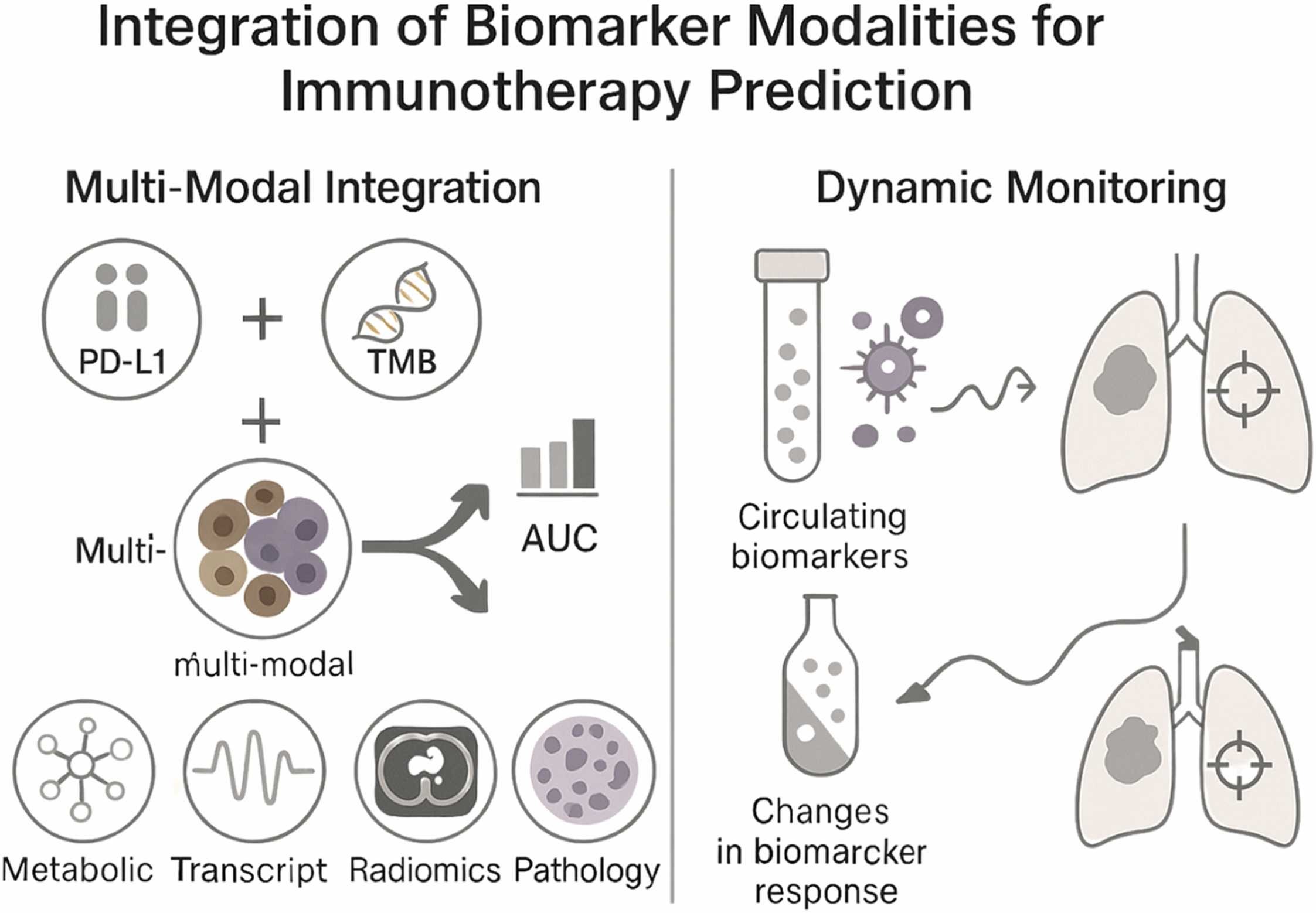
The authors highlight that predicting immunotherapy response cannot rely on any single molecular marker. Instead, multi-parametric models—those integrating molecular, immunologic, and spatial data—are needed to capture the full biological context of the tumor microenvironment (TME).
Beyond genomics, metabolic reprogramming has emerged as a critical determinant of immune evasion and treatment resistance.
Tumors with elevated expression of glucose transporters GLUT1 and GLUT3 exhibit enhanced glycolysis, creating an acidic microenvironment that suppresses T-cell activity. This glucose competition between tumor and immune cells not only limits immune effector function but also promotes immune exhaustion.
Incorporating these metabolic biomarkers into predictive models could refine patient stratification—particularly for tumors that are both metabolically active and immunologically “cold.” Such integrated models could help identify patients who may benefit from therapies that target both metabolism and immune regulation.
Artificial intelligence (AI) and machine learning (ML) now represent the fastest-growing frontiers in predictive oncology. These approaches are capable of integrating high-dimensional clinical, molecular, and imaging data to uncover complex patterns not visible to human observers.
Despite these advances, one major challenge persists: external validation. Many AI models perform exceptionally well within the institution where they were developed but fail to maintain accuracy when tested on independent patient populations—a problem the authors refer to as the “validation gap.”
Combining multiple types of data—genomic, spatial, clinical, and metabolic—has proven far more effective than using single-modality biomarkers.
These multi-modal frameworks have achieved AUC values above 0.85 in several cancers, outperforming traditional metrics. For example, integrating PD-L1 expression, TMB, and immune cell infiltration patterns improved predictive power in non–small cell lung cancer and melanoma.
Modern spatial profiling technologies, such as multiplex immunofluorescence and digital spatial transcriptomics, now reveal how immune and tumor cells are organized within the TME. This spatial information often correlates more strongly with treatment response than bulk biomarker measurements, underscoring the importance of tumor architecture in predicting therapeutic outcomes.
A new generation of mathematical and systems biology models aims to simulate tumor–immune interactions in real time. These models capture key parameters—such as tumor growth kinetics, immune infiltration, and checkpoint blockade dynamics—to forecast treatment outcomes and understand resistance mechanisms.
Early studies show promising results: some of these mechanistic models can classify responders versus non-responders with up to 81% accuracy in pilot validation cohorts. Although still in early stages, these models complement data-driven AI approaches by offering a mechanistic understanding of immune dynamics, which could ultimately guide personalized dosing, combination strategies, and treatment adaptation.
A novel section of the review explores the integration of natural bioactive compounds—such as thymoquinone (Nigella sativa), cucurbitacins (Cucurbitaceae), and organosulfur compounds (garlic derivatives)—which have demonstrated immunomodulatory and metabolic reprogramming effects in preclinical studies. These agents may augment ICI efficacy by:
Such multi-target actions highlight a potential role for metabolic–immune combinatorial therapy, though clinical validation remains limited.
The review identifies three persistent barriers that hinder translation from research to practice:
The authors call for international standardisation frameworks, similar to those of the Global Alliance for Genomics and Health (GA4GH), to harmonise data collection, model validation, and AI governance in oncology.
This landmark review represents one of the most comprehensive syntheses of predictive model development in immuno-oncology. It bridges traditional pathology and next-generation computational science, highlighting the need for multidisciplinary collaboration among pathologists, oncologists, data scientists, and regulatory bodies.
By systematically evaluating every major class of predictive approach—from PD-L1 scoring to AI integration—this work outlines a roadmap for clinically implementable, validated, and interpretable models capable of guiding patient selection and optimizing immune checkpoint inhibitor therapy.
You Can Read Full Article Here